The problem
Utah arrays in primary visual cortex, dual challenges. Dense cortical structure, impedance suited to multi-units recording. Hard to obtain single unit activity (SUA), especially over time. Even with high throughput template matching spike sorters like Kilosort, not trivial to get SUA from large arrays. Experimented with Kilosort 3, seeking better SUA. No magic parameters, but useful tips follow.
Solution part one
First, install Kilosort 3 following the instructions.
- Generate a channel map to linearize Utah array. Geometry not critical; contact sites distant:
Nchannels = 128;
connected = true(Nchannels, 1);
chanMap = 1:Nchannels;
chanMap0ind = chanMap - 1;
xcoords = ones(Nchannels,1)*1000;
ycoords = [1:Nchannels]'*1000;
kcoords = [1:Nchannels]'; % grouping of channels (i.e. tetrode groups)
fs = 30000; % sampling frequency
save('./linear_utah_array.mat', ...
'chanMap','connected', 'xcoords', 'ycoords', 'kcoords', 'chanMap0ind', 'fs')
2) Turn off drift correction in "main_kilosort3.m" - useless in the given geometry :
```matlab
ops.nblocks = 0; % blocks for registration. 0 turns it off, 1 does rigid registration. Replaces "datashift" optio
These two steps should already improve Kilosort 3’s ability to get SUA on Utah arrays.
Soltuion part two
Examine parameter roles when searching for SUAs. Kilosort’s code allows parameter scanning. Rewrite main_kilosort3.m:
function main_ks3_scan(ops)
% Add paths
addpath(genpath('your_path_to_ks_folder')) % path to kilosort folder
addpath('your_path_to_npy_matlab') % for converting to Phy
% Set paths
rootZ = '.\'; % the raw data binary file is in this folder
rootH = '.\'; % path to temporary binary file (same size as data, should be on fast SSD)
pathToYourConfigFile = '.\'; % take from Github folder and put it somewhere else (together with the master_file)
chanMapFile = 'linear_utah_array.mat';
ops.trange = [0 Inf]; % time range to sort
ops.NchanTOT = 128; % total number of channels in your recording
ops.fproc = fullfile(rootH, 'temp_wh.dat'); % proc file on a fast SSD
ops.chanMap = fullfile(pathToYourConfigFile, chanMapFile);
% main parameter changes from Kilosort2 to v2.5
ops.sig = 20; % spatial smoothness constant for registration
ops.fshigh = 300; % high-pass more aggresively
ops.nblocks = 0; % blocks for registration. 0 turns it off, 1 does rigid registration. Replaces "datashift" option.
% Print data location
fprintf('Looking for data inside %s \n', rootZ)
% Check for channel map file
fs = dir(fullfile(rootZ, 'chan*.mat'));
if ~isempty(fs)
ops.chanMap = fullfile(rootZ, fs(1).name);
end
% Find the binary file
fs = [dir(fullfile(rootZ, '*.bin')) dir(fullfile(rootZ, '*.dat'))];
ops.fbinary = fullfile(rootZ, fs(1).name);
% Run the algorithm steps
rez = preprocessDataSub(ops);
rez = datashift2(rez, 1);
[rez, st3, tF] = extract_spikes(rez);
rez = template_learning(rez, tF, st3);
[rez, st3, tF] = trackAndSort(rez);
rez = final_clustering(rez, tF, st3);
rez = find_merges(rez, 1);
% Save results to Phy
rootZ = fullfile(rootZ, 'kilosort3');
mkdir(rootZ)
rezToPhy2(rez, rootZ);
end
Similarly, rewrite StandardConfig_MOVEME file into a function, then write a new MATLAB script:
% Load the default parameters from StandardConfig_MOVEME.m
ops = StandardConfig_MOVEME();
% Define the range of values for the lam, ThPre, and Th parameters
lam_vals = [5, 10, 20, 50];
ThPre_vals = [6, 8, 10, 12];
Th_vals = [8 4; 10 4; 12 4; 14 4];
% Initialize a structure to store the results
% Initialize the result index
result_idx = 1;
% Perform the parameter sweep
for lam_idx = 1:length(lam_vals)
for ThPre_idx = 1:length(ThPre_vals)
for Th_idx = 1:size(Th_vals, 1)
% Update the lam, ThPre, and Th parameter values
ops.lam = lam_vals(lam_idx);
ops.ThPre = ThPre_vals(ThPre_idx);
ops.Th = Th_vals(Th_idx, :);
% Run the mainFunction with the updated ops and store the results
main_ks3_scan(ops);
fprintf('lam: %d, ThPre: %d, Th: [%d, %d]\n', ops.lam, ops.ThPre, ops.Th(1), ops.Th(2));
fprintf('--------------');
end
end
end
Yields several good clusters relative to spike-sorting parameters. However, more clusters not always better. Maximum clusters often result from low stringency thresholds, yielding poorly clustered units with few spikes. Low lam parameter leads to many low amplitude (artifactual?) clusters.
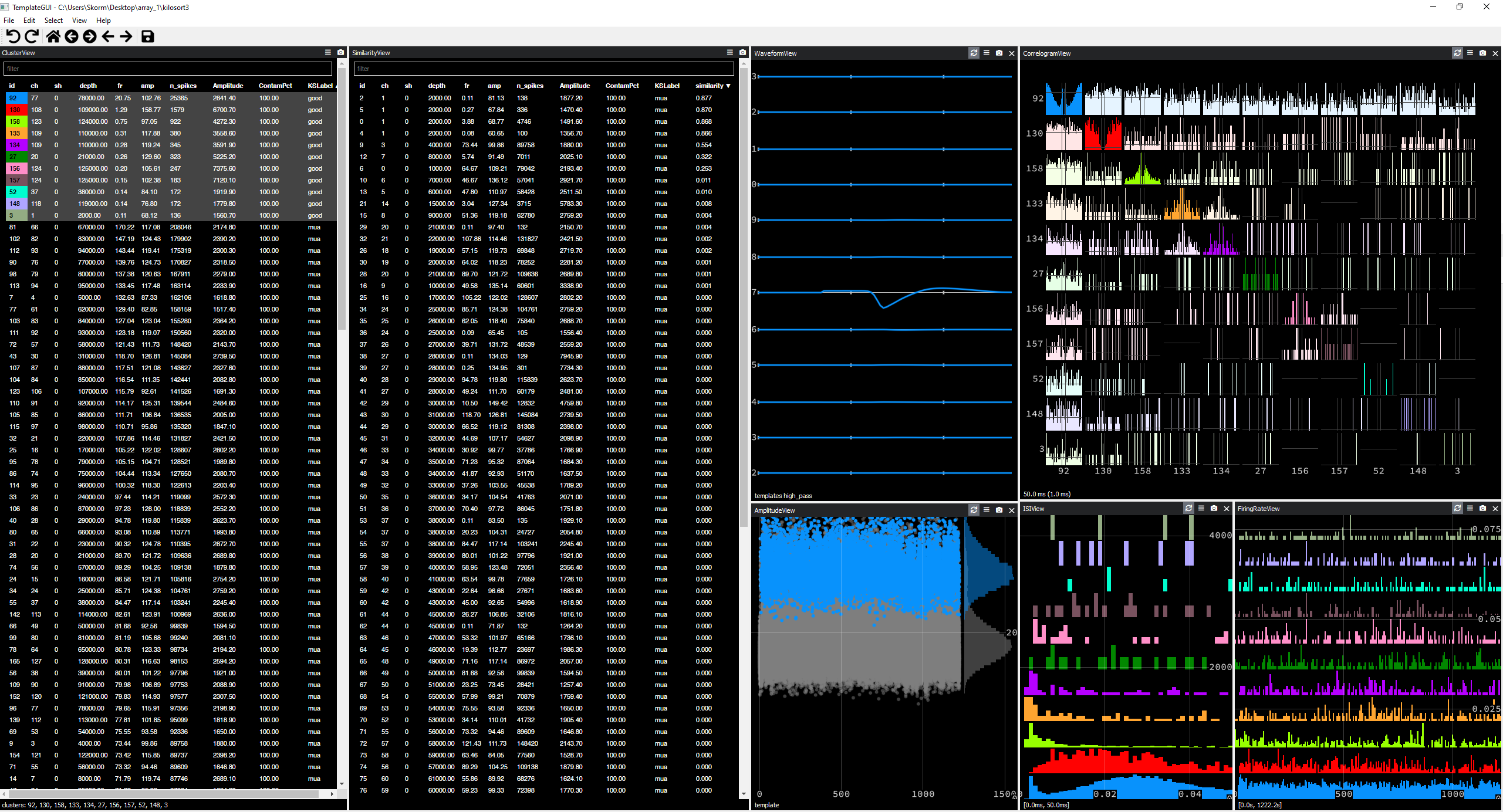
By manually visualizing the clusters with Phy, settled with parameters lam = 10 ; ThPre = 10 ; Th = [12 6], which yields nicer single units :
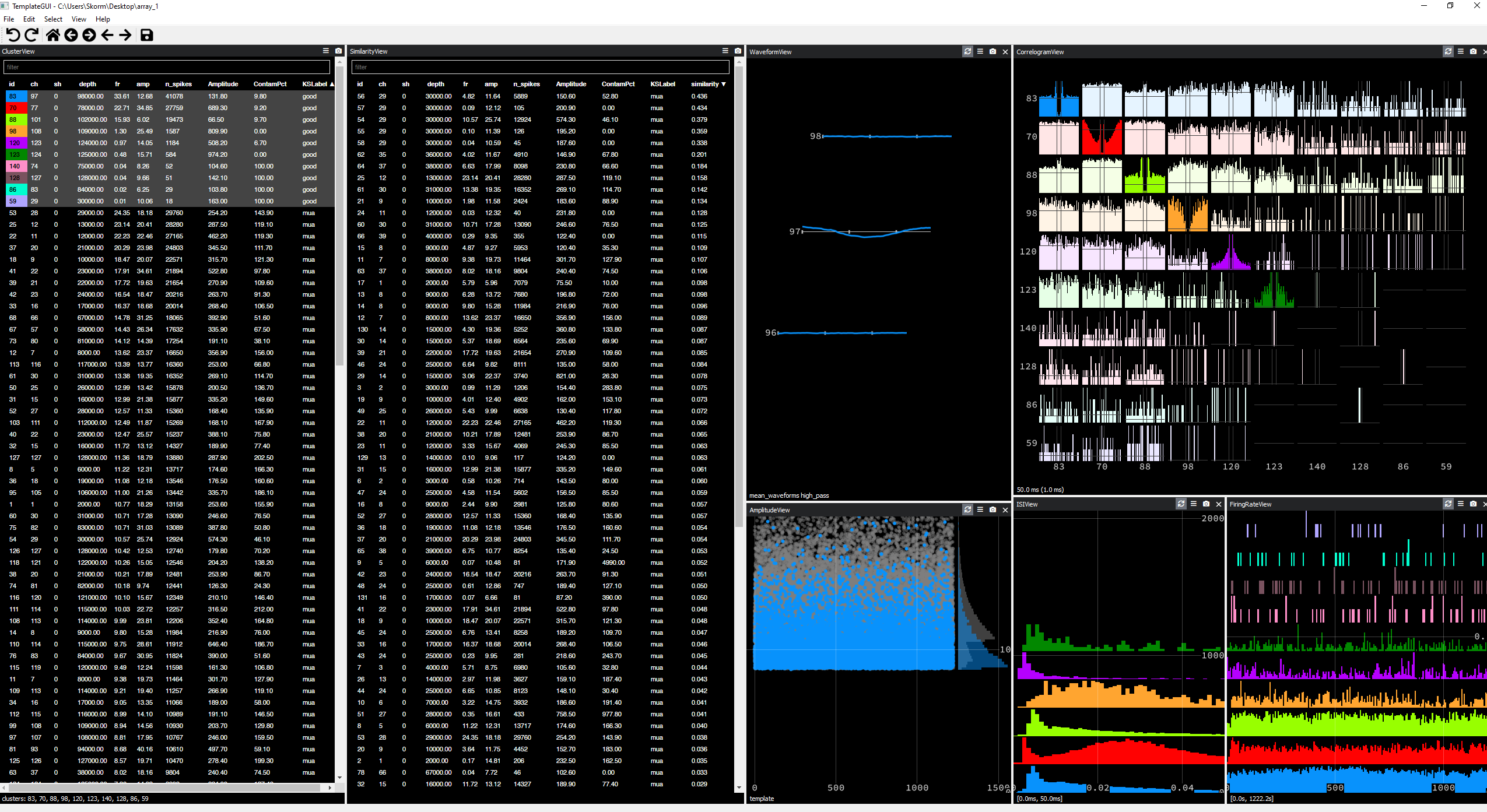
Not a miracle solution, but offers similar performance to Kilosort 2, better at tracking MUAs than KS2.
If miracle solution shows up, will post updates!
